mirror of
https://github.com/infiniflow/ragflow.git
synced 2025-12-26 00:46:52 +08:00
Docs: Updated dataset configuration, KG building and RAPTOR building for v0.21.0 (#10584)
### What problem does this PR solve? ### Type of change - [x] Documentation Update
This commit is contained in:
@ -37,7 +37,7 @@ This section covers the following topics:
|
||||
|
||||
### Select chunking method
|
||||
|
||||
RAGFlow offers multiple chunking template to facilitate chunking files of different layouts and ensure semantic integrity. In **Chunking method**, you can choose the default template that suits the layouts and formats of your files. The following table shows the descriptions and the compatible file formats of each supported chunk template:
|
||||
RAGFlow offers multiple built-in chunking template to facilitate chunking files of different layouts and ensure semantic integrity. From the **Built-in** chunking method dropdown under **Parse type**, you can choose the default template that suits the layouts and formats of your files. The following table shows the descriptions and the compatible file formats of each supported chunk template:
|
||||
|
||||
| **Template** | Description | File format |
|
||||
|--------------|-----------------------------------------------------------------------|-----------------------------------------------------------------------------------------------|
|
||||
@ -54,9 +54,26 @@ RAGFlow offers multiple chunking template to facilitate chunking files of differ
|
||||
| One | Each document is chunked in its entirety (as one). | DOCX, XLSX, XLS (Excel 97-2003), PDF, TXT |
|
||||
| Tag | The dataset functions as a tag set for the others. | XLSX, CSV/TXT |
|
||||
|
||||
You can also change a file's chunking method on the **Datasets** page.
|
||||
You can also change a file's chunking method on the **Files** page.
|
||||
|
||||
|
||||
|
||||
|
||||
:::tip NOTE
|
||||
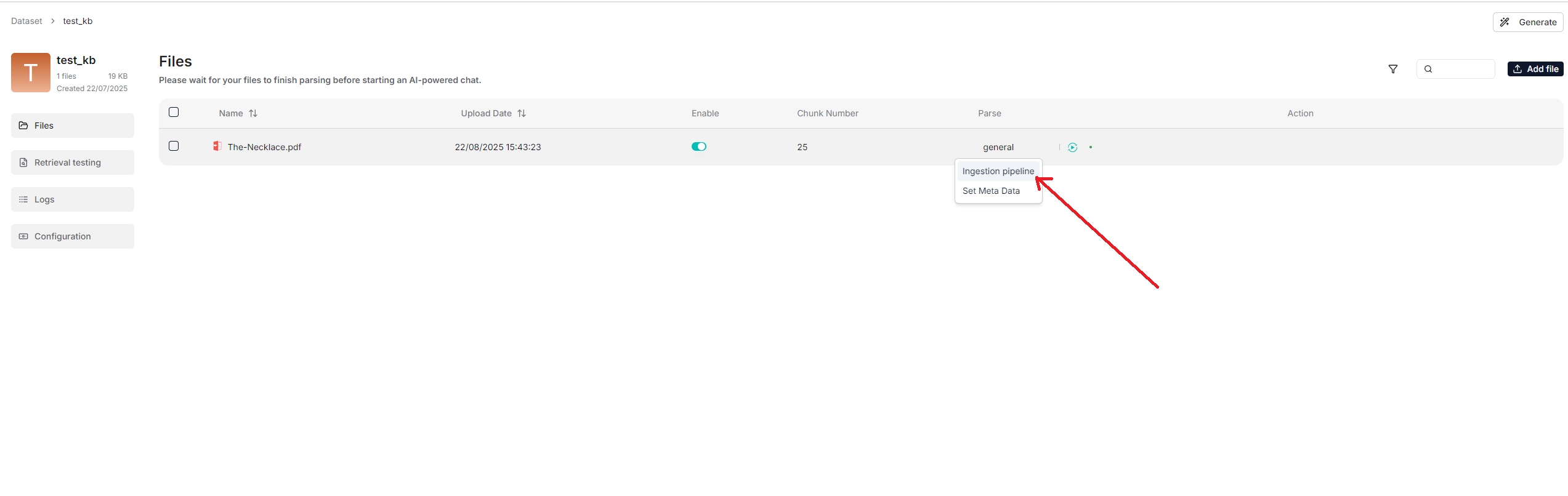
From v0.21.0, RAGFlow supports ingestion pipeline to allow for customized
|
||||
|
||||
<details>
|
||||
<summary>From v0.21.0 onward, RAGFlow supports ingestion pipeline to allow for customized data ingestion and cleansing workflows.</summary>
|
||||
|
||||
To use a customized data pipeline:
|
||||
|
||||
1. On the **Agent** page, click **+ Create agent** > **Create from blank**.
|
||||
2. Select **Ingestion pipeline** and name your data pipeline in the popup, then click **Save** to show the data pipeline canvas.
|
||||
3. After updating your data pipeline, click **Save** on the top right of the canvas.
|
||||
4. Navigate to the **Configuration** page of your dataset, select **Choose pipeline** in **Ingestion pipeline**.
|
||||
|
||||
*Your saved data pipeline will appear in the dropdown menu below.*
|
||||
|
||||
</details>
|
||||
|
||||
### Select embedding model
|
||||
|
||||
|
||||
Reference in New Issue
Block a user